Other phylogenies


Other phylogenies




Back to the
“MOLECULAR PHYLOGENETICS” page
Back to the
“LANE”
introduction page

FACTS
Taxonomic groups:
‣Myzostomes
‣Chrysomelid beetles
‣Toxin-Antitoxin in bacteria
‣Rotifers

PUBLICATIONS
‣Systematic Biology 46: 722-747 (1997)
‣Proceedings of the Royal Society, London, B, 267: 1383-1392 (2000)
‣PNAS, 98: 10202-10207 (2001)
‣PNAS, 98: 3909-3914 (2001)
‣Proceedings of the Royal Society, London, B, 269: 1-6 (2002)
‣Molecular Phylogenetics & Evolution 31: 1–3 (2004)
‣Biotechniques 38: 775-781 (2005)
‣Systematic Biology 55: 208–227 (2006)
‣Molecular Biology & Evolution 24(8): 1690–1701 (2007)
‣Biological reviews 82 (4): 551-572 (2007)
‣BMC Microbiology, 8: 104 (2008)
‣PloS Genetics (in press)

Related publications from our group on the phylogeny of other groups
‣Check the main “Molecular Phylogenetics” page

PEOPLE INVOLVED
FROM MICHEL
MILINKOVITCH’S LAB
‣Arnaud Termonia
‣Patrick Mardulyn
‣Julien Guglielmini

MAIN COLLABORATORS
Jacques Pasteels
Has been Full Professor at the Université Libre de Bruxelles, and is now Emeritus Professor at the same university
Igor Eeckhaut
Is Professor in the Laboratory of Marine Biology at the University of Mons-Hainaut (Belgium)
Ralph Tiedemann
Was a Post-doc in Michel Milinkovitch’s lab. He is now Full Professor at the University of Potsdam, Germany.
Karin Van Doninck
Was a Post-doc in Matthew Meselson’s lab. She is now Assistand Professor at the University of Namur, Belgium.



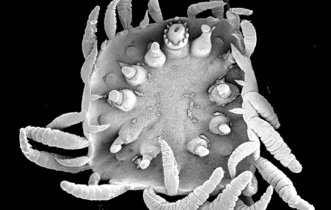
Molecular data opens the massive realms of living species to phylogenetic scrutiny
Evolutionary concepts are pertinent not only to that large diversity of possible applications across disciplines, but also to the massive realm of diversity across living beings - i.e., from natural/artificial clones to the major kingdoms made of approximately 50 millions of extant species, all living beings are connected through pedigrees and the phylogenetic tree of life. If horizontal transfer (including hybridization) and differential lineage sorting are rare phenomena (which is clearly not always the case for prokaryotes which experience significant horizontal transfer), gene trees are estimates of the phylogeny among the species bearing these genes.
Although, in the molecular phylogenetic field, we have mostly focused on cetaceans and ranid frogs, we have also used molecular phylogenetic approaches for studying the phylogeny of a variety of other groups all over the tree of life. Molecular epidemiology examples are available HERE, and we present below our work on the phylogeny of chrysomelid beetles (insects), myzostomes, and rotifer lineages.
(1) Insects: a combination of host-plant affiliation, spectrometry analysis of defensive secretions, and molecular phylogenetics
✓Termonia A., Hsiao T. H., Pasteels J.M. & M. C. Milinkovitch
Feeding specialization and host-derived chemical defense in Chrysomeline leaf beetles did not lead to an evolutionary dead end
PNAS, 98: 3909-3914 (2001)
➡ABSTRACT. Combination of molecular phylogenetic analyses of Chrysomelina beetles and chemical data of their defensive secretions indicate that two lineages independently developed, from an ancestral autogenous metabolism, an energetically efficient strategy that made the insect tightly dependent on the chemistry of the host plant. However, a lineage (the interrupta group) escaped this subordination through the development of a yet more derived mixed metabolism potentially compatible with a large number of new host-plant associations. Hence, these analyses on leaf beetles document a mechanism that can explain why high levels of specialization do not necessarily lead to ‘‘evolutionary dead ends.’’
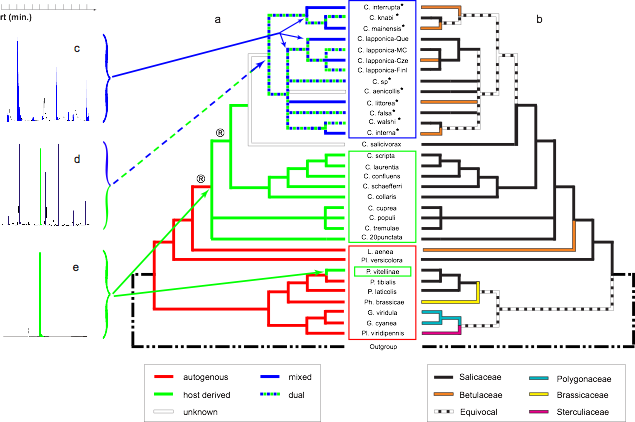
➡Associated publications:
✓Mardulyn P., Milinkovitch M. C., & J. M. Pasteels
Phylogenetic Analyses of DNA and Allozyme Data Suggest that Gonioctena Leaf Beetles (Coleoptera; Chrysomelidae) Experienced Convergent Evolution in Their History of Host-Plant Family Shifts
Systematic Biology 46: 722-747 (1997)
✓Termonia A., Pasteels J.M., Windsor D.M. & M. C. Milinkovitch
Dual Chemical Sequestration, a Key Mechanism in Transitions Among Ecological Specializations
Proceedings of the Royal Society, London, B, 269: 1-6 (2002)

✓Eeckhaut I., McHugh D., Mardulyn P., Tiedemann R., Monteyne D., Jangoux M., & M C. Milinkovitch
Myzostomida: a Link between Trochozoans and Flatworms
Proceedings of the Royal Society, London, B, 267: 1383-1392 (2000)
Cited as «A puzzling Metazoan Body Plan», Editor’s Choice Section of Science, 289: 217
✓Bleidorn C., Eeckhaut I., Podsiadlowski L., Schult N., Mchugh D., Halanych K.M., Milinkovitch M. C. & R. Tiedemann
Mitochondrial genome and nuclear sequence data support Myzostomida as part of the annelid radiation
Molecular Biology & Evolution 24(8): 1690–1701 (2007)
➡ABSTRACT. The echinoderm symbionts Myzostomida are marine worms that show an enigmatic lophotrochozoan body plan. Historically, their phylogenetic origins were obscured due to disagreement about which morphological features are evolutionarily conserved, but now most morphological evidence points to annelid origins. In contrast, recent phylogenetic analyses using different molecular markers produced variable results regarding the position of myzostomids, but all suggested these worms are not derived annelids. To reexamine this issue, we analyzed data from nuclear genes (18S rDNA, 28S rDNA, Myosin II, and Elongation Factor-1a), and a nearly complete myzostomid mitochondrial genome. Here, we show that the molecular data are in agreement with the morphological evidence that myzostomids are part of the annelid radiation. This result is robustly supported by mitochondrial (gene order and sequence data) and nuclear data, as well as by recent ultrastructural investigations. Using Bayes factor comparison, alternative hypotheses are shown to lack support. Thus, myzostomids probably evolved from a segmented ancestor and gained a derived anatomy during their long evolutionary history as echinoderm symbionts.
➡REMARK. These alien organisms are extremely divergent morphologically. They probably belong to one of the first offshoot within annelids.
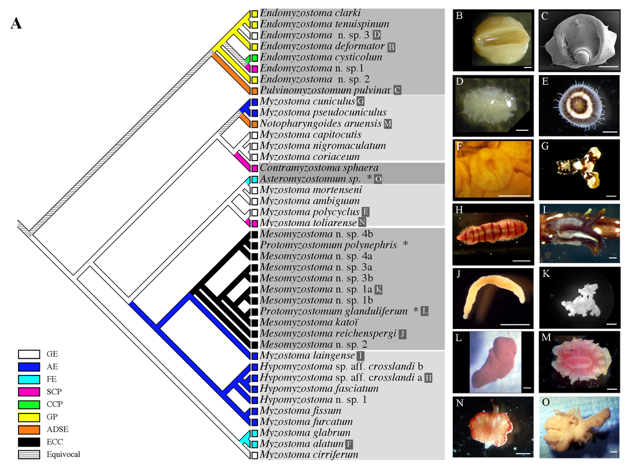
(2b) Myzostomes evolved parasitism multiple times
✓Lanterbecq D., Rouse G.W., Milinkovitch M.C. & I. Eeckhaut
Molecular Phylogenetic Analyses Indicate Multiple Independent Emergences of Parasitism in Myzostomida (Protostomia)
Systematic Biology 55: 208–227 (2006)
➡ABSTRACT. The fossil record indicates that Myzostomida, an enigmatic group of marine worms, traditionally considered as annelids, have exhibited a symbiotic relationship with echinoderms, especially crinoids, for nearly 350 million years. All known extant myzostomids are associated with echinoderms and infest their integument, gonads, celom, or digestive system. Using nuclear (18S rDNA) and mitochondrial (16S and COI) DNA sequence data from 37 myzostomid species representing nine genera, we report here the first molecular phylogeny of the Myzostomida and investigate the evolution of their various symbiotic associations. Our analyses indicate that the two orders Proboscidea and Pharyngidea do not constitute natural groupings. Character reconstruction analyses strongly suggest that (1) the ancestor of all extant myzostomids was an ectocommensal that first infested crinoids, and then asteroids and ophiuroids, and (2) parasitism in myzostomids emerged multiple times independently.
(3) The phylogeny of bacterial toxins (from TA systems).
Toxin-Antitoxin systems are plasmid spectacular maintenance mechanisms that make them purely "selfish DNA" candidates. A TA system is made of a gene coding for a cytotoxic stable protein preceded by a gene coding for an unstable antitoxin. The toxin being more stable than the antitoxin, absence of the operon causes a reduction of the amount of the latter relative to the amount of the former. Thus, a cell exhibiting a TA system on a plasmid is 'condemned' either not to loose it or to die. In the context of our phylogenetic analyses of TA systems, we developed an algorithm that searches public databases on the basis of predefined similarity and TA-specific structural constraints. This approach, using a single starting query sequence for each of the TA families identified over 1,500 putative TA systems. These groups of sequences were analyzed phylogenetically for a better classification and understanding of TA systems evolution. The resulting phylogenetic trees are available for browsing and searching through a small java program (TIQ) available HERE. Much additional information is available in our full publication:
✓Guglielmini J., Szpirer C., & M. C. Milinkovitch
Automated Discovery and Phylogenetic Analysis of New Toxin-Antitoxin Systems
BMC Microbiology, 8: 104 (2008)
Note that TA systems have been domesticated as biotechnological tools for facilitating DNA engineering and protein production without the use of antibiotics:
✓Szpirer C. Y. & M. C. Milinkovitch
Separate-Component-Stabilization (SCS) System for protein and DNA production without the use of antibiotics
Biotechniques 38: 775-781 (2005)
The company Delphi Genetics develops and commercialise technologies and tools for facilitating DNA engineering (see our “Applied Evolutionary Genetics” web page).
(4) Rotifers have evolved unique histone variants possibly associated to their ability to repair massive levels of DNA breakage
✓Van Doninck K., Mandigo M.L., Hur J.H., Wang P., Guglielmini J., Milinkovitch M.C., Lane W.S. & M. Meselson
Phylogenomics of Unusual Histone H2A Variants in Bdelloid Rotifers
PloS Genetics PloS Genetics 5 (3): e1000401
➡ABSTRACT. Rotifers of Class Bdelloidea are remarkable in having evolved for millions of years apparently without males and meiosis. In addition, they are unusually resistant to desiccation and ionizing radiation and are able to repair hundreds of radiation-induced DNA double strand breaks per genome with little effect on viability or reproduction. As specific histone H2A variants are involved in DSB repair and certain meiotic processes in other eukaryotes, we investigated the histone H2A genes and proteins of two bdelloid species. Genomic libraries were built and probed to identify histone H2A genes in the genomes of Adineta vaga and Philodina roseola, species representing two different bdelloid families. The expressed H2A proteins were visualized on SDS-PAGE gels and identified by tandem mass spectrometry. We find that neither the core histone H2A, present in nearly all other eukaryotes, nor the H2AX variant, a ubiquitous component of the eukaryotic DSB repair machinery, are present in bdelloid rotifers. Instead, they are replaced by unusual histone H2A variants of higher mass. In contrast, a species of rotifer belonging to the facultatively sexual, desiccation and radiation-intolerant sister class of bdelloid rotifers, the monogononts, contains a canonical core histone H2A and appears to lack the bdelloid H2A variant genes. Applying phylogenetic tools we demonstrate that the bdelloid-specific H2A variants arose as distinct lineages from canonical H2As and are not related to the universally conserved H2AX or H2AZ variants. The replacement of core H2A and H2AX in bdelloid rotifers by previously uncharacterized H2A variants with extended carboxy-terminal tails indicates evolutionary diversity within this class of histone H2A genes and may represent adaptation to unusual features specific to bdelloid rotifers
➡REMARK. Bdelloid rotifers are microscopic animals common in freshwater and temporary environments throughout the world. They are very peculiar organisms not only because they have been reproducing without males for millions of years, but also because they can survive high levels of radiations and long periods of complete desiccation, despite that these two phenomena shatter DNA into pieces. Canonical histones (H2A, H2B, H3 and H4) are very conserved proteins that package DNA in the nucleus and regulate the chromatin metabolism. One evolutionary conserved histone variant of canonical H2A, H2AX, is involved in repairing DNA double strand breaks. We therefore expected to find a high amount of H2AX in bdelloid rotifers. Strikingly, we find that bdelloids lack both H2A and H2AX histones. The absence of these two conserved proteins is in remarkable contrast to their ubiquitous presence in other eukaryotes. Instead, we find that bdelloid rotifers replaced their canonical H2A protein by unique H2A variants not found in any other eukaryote. These results gain particular interest when extreme resistance of bdelloid rotifers to desiccation and ionizing radiations is taken into account and their attendant ability, possibly unique among metazoans, to repair massive levels of DNA breakage.
(5) Others

✓Milinkovitch M. C., Caccone A. & G. Amato
Molecular phylogenetic analyses indicate extensive morphological convergence between the ‘‘yeti’’ and primates (April’s fool day paper in the 1st of April Issue of MPE)
Molecular Phylogenetics and Evolution 31: 1–3 (2004)
➡REMARK. This is an April’s fool day paper ... but it tuned out to be very popular, especially for teaching students about forensics and phylogeny. Read the full story HERE.
✓Levasseur A., Orlando L., Bailly X, Milinkovitch M.C., Danchin E. & P. Pontarotti
Criteria for quantifying the role of environmental changes on gen(om)e evolution: the roles of positive selection and neutral evolution
Biological reviews 82 (4): 551-572 (2007)
