Phylogeny of cetaceans


Phylogeny of cetaceans




Back to the
“MOLECULAR PHYLOGENETICS” page
Back to the
“LANE”
introduction page

FACTS
Species:
‣Whales, dolphins, porpoises, ... and artiodactyls.

PUBLICATIONS
‣Book chapter: p317-338 in ‘Molecular Genetics of Marine Mammals’, Special Publication Number 3; The Society for Marine Mammology (1998)
‣Book chapter: p113-131 in ‘The Emergence of Whales: Evolutionary Patterns in the Origin of Cetacea’, Plenum, New York 1998
‣Book chapter: p325-359 in ‘Marine Mammals: Biology and Conservation’, Kluwer Academic Press, 2002
‣Journal of Evolutionary Biology, 5: 149-160 (1992)
‣Nature, 361: 346-348 (1993)
‣Marine Mammal Science, 10: 125-128 (1994)
‣Molecular Biology & Evolution, 11: 939-948 (1994)
‣Molecular Biology & Evolution, 12: 518-520 (1995)
‣Trends in Ecology and Evolution, 10: 328-334 (1995)
‣Genetics, 144: 1817-1833 (1996)
‣Journal of Molecular Evolution, 44(Suppl 1): S117-S120 (1997)
‣Nature, 388: 622-624 (1997)
‣Systematic Biology, 48: 6-20 (1999)
‣Molecular Phylogenetics and Evolution, 15(2): 314–318 (2000)
‣PNAS, 97: 11343-11347 (2000)
‣Systematic Biology, 49: 808-817 (2000)
‣Science (Letter), 294, 787 (2001)

Related publications from our group on the population & conservation genetics of cetaceans
‣Check the main “Conservation Genetics” page

PEOPLE INVOLVED
FROM MICHEL
MILINKOVITCH’S LAB
‣Insa Cassens
‣Athanasia Tzika
‣Sabrina Rosa
‣Daniel Monteyne

MAIN COLLABORATORS
John Gatesy
Assistant Professor at University of California Riverside, USA
Jeff Powell
Professor at Yale University, USA
Axel Meyer
Professor at the University of Konstanz, Germany
Masami Hasegawa
He was Professor at the Institute of Statistical Mathematics, Tokyo, Japan, and he is now Professor at Fudan University, China.
Michael Stanhope
Professor at Cornell University, USA
Rick LeDuc & Andrew Dizon
Southwest Fisheries Science Center, California, USA




Cetacean phylogeny, a difficult problem
Because cetaceans experienced dramatic transformations in basically all their biological systems, phylogeneticists encountered obvious difficulties in assessing homology and polarisation of the few morphological characters that could be used for comparisons between cetaceans and other mammals. Despite these difficulties, morphological analyses have suggested that cetaceans are closely related to artiodactyls. While morphologists and molecular phylogeneticists heatedly argued over several other nodes of the mammalian phylogenetic tree, the morphological hypothesis of a close relationship between cetaceans and ungulates (and especially artiodactyls) was strongly supported by a range of molecular data: from immunological (e.g., Boyden and Gemeroy, 1950; Shoshani, 1986) and DNA-DNA hybridisation (Milinkovitch, 1992) studies, to analyses of mitochondrial DNA (mtDNA) and nuclear amino-acid and DNA sequences (e.g., Czelusniak et al., 1990; Milinkovitch et al., 1993). This is rather a counter-intuitive result because it makes a cow, a pig, or a camel more closely related to a dolphin or a blue whale than to a horse or a tapir. However, Graur and Higgins (1994) analysed a large data set of published amino-acid and DNA sequences and first suggested explicitly an even more provocative hypothesis: cetaceans would be nested within the artiodactyl phylogenetic tree.
This hypothesis of artiodactyl paraphyly has subsequently been supported by (among others):
1.Extensive cladistic analyses of DNA sequence data from multiple nuclear and mitochondrial genes (Gatesy, 1998);
2.Lactalbumin DNA sequences (Milinkovitch et al., 1998),
3.Phylogenetic interpretation of retropositional events that lead to the insertion of short interspersed elements (SINEs) at particular loci in the nuclear genome of various artiodactyl and cetacean ancestors (Shimamura et al., 1997; Milinkovitch and Thewissen, 1997).
Extensive phylogenetic re-analyses (Gatesy et al., 1999) of all available data indicate that the inclusion of a monophyletic Cetacea within the phylogenetic tree of Artiodactyla has been stable to increased taxonomic sampling and to the addition of nearly nine thousand characters from three mtDNA genes, 12 nuclear genes, and eight SINE retroposon loci. Although the concept of cetaceans as highly derived artiodactyls still raises major scepticism (e.g., Luckett and Hong, 1998; Heyning, 1999), it constitutes one of the best supported hypotheses of interordinal (sensu lato) relationship within the phylogeny of mammals.
Similarly, the phylogenetic relationships among the major lineages of cetaceans has generated much controversies. These web pages do NOT constitute an attempt to review the problem but simply report on our publications on the topic. These publications are too numerous to be detailed here. We simply provide the reference of the paper, the abstract, and a short remark to put the study in context. Please, consult the full publications for references and much additional information.
(1) The origin of cetaceans within mammals
✓Milinkovitch M. C.
DNA-DNA Hybridizations Support Ungulate Ancestry of Cetacea
Journal of Evolutionary Biology, 5: 149-160 (1992)
➡ABSTRACT. Recent morphological data on Pakicetus spp. and Basifosaurus spp. fossils suggest that cetaceans (whales, dolphins and porpoises) originate from carnivorous Mesonychid land mammals (Condylarthra) and made a gradual transition from land to sea in early Eocene (Gingerich et al. 1983; 1990). On the other hand, there is convincing evidence that Artiodactyla and Perissodactyla have evolved from Condylarthra (Van Valen 1978, Carrel 1988). Therefore, the Pakicetus and Basilosaurus data suggest a close genetic relationship between cetaceans and ungulates. An approach based on molecular genetics was used in this study to test the morphological hypothesis. Liver samples of two Delphinoidea species were obtained from animals caught in a Peruvian gillnet fishery. 32P- or 35S-labeled single copy nuclear genomes (scn-DNA) of the two cetacean species were hybridized each with unlabeled total DNA of various cetaceans, ungulates and other mammals including primates, rodents, lagomorphs and carnivores. The Tmedian (Tm) and Tmode of all melting curves, used as a measure of the DNA-DNA hybrids stability, clearly show a greater sequence similarity - and thus a lower genetic distance - between cetaceans and ungulates than between cetaceans and other mammals.
➡REMARK. Geeee, this is old. Nobody uses this method anymore (heteroduplex DNA-DNA hybridization in solution) ... but it has the advantage of providing a measure of similarity between two “full” (single-copy) genomes. The method has many disadvantages as well (beside being a pain in the neck). Still, it was one of the very first studies using molecular techniques for estimating the phylogenetic position of cetaceans within mammals.
✓Milinkovitch M. C., Bérubé M. & P. J. Palsbøll
Cetaceans Are Highly Specialized Artiodactyls Pages 113-131 in: ‘The Emergence of Whales: Evolutionary Patterns in the Origin of Cetacea’, (Thewissen Ed.), Plenum, New York 1998.
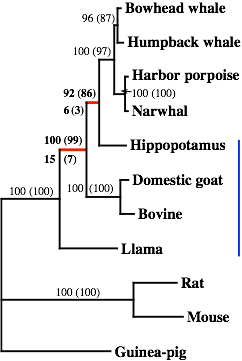
✓Gatesy J., Milinkovitch M. C., Waddelld V. & M. Stanhope
Stability of Cladistic Relationships between Cetacea and Higher Level Artiodactyl Taxa
Systematic Biology, 48: 6-20 (1999)
➡ABSTRACT. Over the past 10 years, the phylogenetic relationships among higher-level artiodactyltaxa have been examined with multiple data sets. Many of these data sets suggest that Artiodactyla (even-toed ungulates) is paraphyletic and that Cetacea (whales) represents a highly derived “artiodactyl” subgroup. In this report, phylogenetic relationships between Cetacea and artiodactyls are tested with a combination of 15 published data sets plus new DNA sequence data from two nuclear loci, interphotoreceptor retinoid-binding protein (IRBP) and von Willebrand factor (vWF). The addition of the IRBP and vWF character sets disrupts none of the relationships supported by recent cladistic analyses of the other 15 data sets. Simultaneous analyses support three critical clades: (Cetacea + Hippopotamidae), (Cetacea + Hippopotamidae + Ruminantia), and (Cetacea + Hippopotamidae + Ruminantia + Suina). Perturbations of the combined matrix show that the above clades are stable to a variety of disturbances. A chronicle of phylogenetic results over the past 3 years suggests that cladistic relationships between Cetacea and artiodactyls have been stable to increased taxonomic sampling and to the addition of more than 1,400 informative characters from 15 data sets.
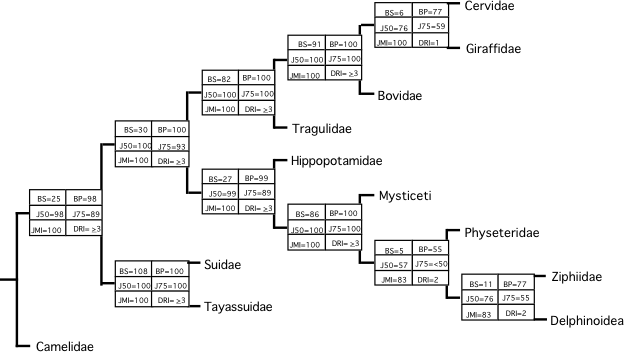
Figure: The shortest topology for one of the super-matrix dataset analysed in this paper. At each node, various support values are indicated.
➡REMARK. This paper demonstrates the stability of the phylogenetic position of cetaceans WITHIN artiodactyls. Large taxon sampling, large number of characters from a variety of loci. Difficult to invoke analytical problems. It makes the “Cetartiodactyla” (cetaceans nested within artiodactyls) clade one of the best supported lineage in the phylogeny of mammals. This result was confirmed by the analysis of insertion events involving SINEs (short interspersed elements) retroposons performed by Okada’s group in Japan (Shimamura et al. Nature 388, 666–670 [1997]). Below is the reference of the News&Views I wrote with Hans Thewissen about Shimamura et al.’s paper. The second reference is a paper where we describe the pros and cons of using SINEs for phylogeny inference.
✓Milinkovitch M. C. & J.G.M. Thewissen
Eventoed Fingerprints on Whale Ancestry
Nature, 388: 622-624 (1997)
✓Shedlock A.M., Milinkovitch M. C. & N. Okada
SINE evolution, missing data, and the origin of whales
Systematic Biology, 49: 808-817 (2000)
(2) The phylogenetic relationships among cetaceans
✓Milinkovitch M. C., Ortí G. & A. Meyer
Revised Phylogeny of Whales Suggested by Mitochondrial Ribosomal DNA Sequences
Nature, 361: 346-348 (1993)
➡ABSTRACT. Living cetaceans are subdivided into two highly distinct suborders, Odontoceti (the echolocating toothed whales) and Mysticeti (the filter-feeding baleen whales), which are believed to have had a long independent history. Here we report the determination of DNA sequences from two mitochondrial ribosomal gene segments (930 base pairs per species) for 16 species of cetaceans, a perissodactyl and a sloth, and construct the first phylogeny for whales and dolphins based on explicit cladistic methods. Our data (and earlier published myoglobin sequences) confirmed that cetaceans are closely related to artiodactyls and that all families and superfamilies of cetaceans are monophyletic. A surprising finding was that one group of toothed whales, the sperm whales, is more closely related to the baleen whales than to other odontocetes. The common ancestor of baleen whales and sperm whales might have lived only 10–15 million years ago. The suggested paraphyly of toothed whales has many implications for classification, phylogeny and our understanding of the evolutionary history of cetaceans.
➡REMARK. The result of a close relationship between cetaceans and artiodactyls was well accepted (and confirmed later, see below) but the sister-relationship between sperm whales and baleen whales (making toothed whales paraphyletic) generated a controversy. Several studies based on mitochondrial DNA sequences supported the paraphyly of toothed whales (see additional references below) ... but this hypothesis has been since contradicted by extended analyses of nuclear sequences. There is today little doubt that toothed whales are monophyletic but the tendency of several datasets (especially mitochondrial) to groups sperm and baleen whales remains unexplained. We still don’t know if this is due to genuine phylogenetic signal (caused by differential lineage sorting) or if it is simply an artifact due to an association of low taxon sampling, short sequences, and immature methodologies (many of us were sinners in these early days of phylogenetics, thinking we could build a phylogeny with a few hundreds nucleotides from a handful species).
➡Other, similar, references:
✓Milinkovitch M. C., Meyer A. & J. R. Powell
Phylogeny of All Major Groups of Cetaceans Based on DNA Sequences from Three Mitochondrial Genes
Molecular Biology & Evolution, 11: 939-948 (1994)
✓Milinkovitch M. C., Ortí G. & A. Meyer
Novel Phylogeny of Whales Revisited but not Revised
Molecular Biology & Evolution, 12: 518-520 (1995)
✓Milinkovitch M.C.
Molecular Phylogeny of Cetaceans Prompts Revision of Morphological transformations
Trends in Ecology and Evolution, 10: 328-334 (1995)
✓Milinkovitch M. C., Leduc R. G., Adachi J., Farnir F., Georges M., & M. Hasegawa
Effects of character weighting and species sampling on phylogeny reconstruction: a case study based on DNA sequence data in cetaceans
Genetics, 144: 1817-1833 (1996)
✓Hasegawa M., Adachi J., & M. C. Milinkovitch
Novel Phylogeny of Whales Supported by Total Molecular Evidence
Journal of Molecular Evolution, 44(Suppl 1): S117-S120 (1997)
✓Milinkovitch M. C.
The Phylogeny of Whales: a Molecular Approach. Pages 317-338 in: ‘Molecular Genetics of Marine Mammals’, (A. Dizon et al. Eds.), Special Publication Number 3
The Society for Marine Mammology (1998)
✓Waddell V. G., Milinkovitch M. C., Bérubé M. & M. J. Stanhope
Molecular phylogenetic examination of the Delphinoidea trichotomy: Congruent evidence from three nuclear loci indicates that porpoises (Phocoenidae) share a more recent common ancestry with white whales (Monodontidae) than they do with true dolphins (Delphinidae).
Molecular Phylogenetics and Evolution, 15(2): 314–318 (2000)
➡ABSTRACT. Porpoises (Phocoenidae), dolphins (Delphinidae), and the two species of Monodontidae (beluga and narwhal) together constitute the superfamily Delphinoidea. Although there is extensive evidence supporting the monophyly of this superfamily, previous studies involving morphology, as well as sequence analysis of mitochondrial genes, have failed to yield a clear picture of the relative relationships within the group. Here we present the first examination of this issue from the perspective of single-copy nuclear genes at the DNA sequence level. The data involve three such loci: von Willebrand factor (vWF), interphotoreceptor retinoid binding protein (IRBP), and lactalbumin. The vWF and IRBP data sets consist of protein-coding fragments, whereas the sequenced lactalbumin fragment is predominately intronic. All phylogenetic analyses involving at least one representative from each of the three Delphinoidea families congruently support a beluga/porpoise clade. The levels of sequence divergence for most of these data appear to roughly concur with a paleontological date for the radiation of the Delphinoidea at 11–15 MYA but, in agreement with mitochondrial DNA sequence analyses, suggest that the extant major groups of cetaceans radiated approximately 25 MYA, 10 million years later than inferred from paleontological data.
➡REMARK. Abstract is self-explanatory. This is the first study showing the sister relationship between porpoises (Phocoenidae) and white whales (Monodontidae: the beluga and the narwhal).
✓Milinkovitch M. C., LeDuc R., Tiedemann R., & A. Dizon
Applications of Molecular Data in Cetacean Taxonomy and Population Genetics with Special Emphasis on Defining Species Boundaries
Pages 325-359 in: ‘Marine Mammals: Biology and Conservation’ (Evans, P. G. H., and J. A. Raga eds.), Kluwer Academic Press, 2002.
➡REMARK. This is a book chapter, so there is no abstract. The main focus of this chapter lies at the junction between studies of the relationships among species and studies of intraspecific population structure. In other words, this work deals with the study of the differentiation of species (alpha taxonomy). It involves the number and names of species in a classification. We discuss current alpha taxonomy in cetaceans, the relative merits of several molecular approaches for identifying species boundaries (lack of shared haplotypes vs. presence of fixed differences vs. reciprocal monophyly vs. degree of divergence among sequences). Finally, to provide a framework for the interpretation and use of genetic data, we briefly examine some chosen case studies, where molecular evidence has already contributed or might potentially contribute to the clarification of species boundaries in cetaceans.
(3) The special case of “river dolphins”
✓Milinkovitch M. C. & I. Cassens
Deciphering River Dolphin Evolution
Science (Letter), 294, 787 (2001)
✓Cassens I., Vicario S., Waddell V. G., Balchowsky H., Van Belle D., Wang Ding, Chen Fan, Lal Mohan R.S., Simões-Lopes P. C., Bastida R., Meyer A., Stanhope M. J. & M. C. Milinkovitch
Independent Adaptation to Riverine Habitats Allowed Survival of Ancient Cetacean Lineages
PNAS, 97: 11343-11347 (2000)
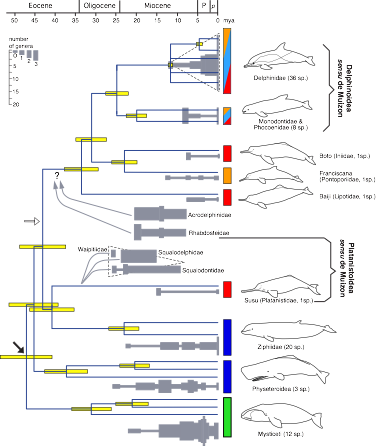
➡REMARK. One of my favorite papers combining molecular phylogeny inference, bayesian approaches to dating nodes in the tree, and paleontological data. We suggest that the so-called river dolphins is a wastebasket polyphyletic taxon consisting of relict dolphin lineages from the Eocene, Oligocene, and early Miocene, that developed ecological extreme specialization (including for at least three of the four lineages, adaptation to the fluvial habitat) which incidentally insured their survival against either major changes of physical parameters in the marine environment or the emergence and radiation of better adapted and more recent marine small cetaceans. Ironically, these river dolphins are now among the most vulnerable species of cetaceans (because of the massive impact of human activities on riverine habitats). The Chinese river dolphin (Lipotes vexillifer) is considered extinct in the wild since 2005.
Others
✓Milinkovitch M. C., Dunn J. L. & J. R. Powell
Exfoliated Cells as the Most Accessible DNA Source for Captive Whales and Dolphins
Marine Mammal Science, 10: 125-128 (1994)





