Phylogeography of Colour Variation
in Panther Chameleons


Phylogeography of Colour Variation
in Panther Chameleons




Back to the
“Conservation Genetics” page
Back to the
“LANE”
introduction page



FACTS
Species:
‣Furcifer pardalis
... but should probably be split into 4 to 11 species
Locations:
‣North of Madagascar

PUBLICATIONS
‣Molecular Ecology (2015)

Related publications from our group on population genetics
‣Check the main “Conservation Genetics” page

PEOPLE INVOLVED
FROM MICHEL
MILINKOVITCH’S LAB
‣Suzanne Saenko
‣Adrien Debry

MAIN COLLABORATORS IN MADAGASCAR
✓Achille Raselimanana

Professor at the University of Antananarivo & President of the Association Vahatra
✓Toky Randriamoria

Student of Prof. Raselimanana at the University of Antananarivo.



Phylogeography and Support Vector Machine Classification of Colour Variation in Panther Chameleons
Lizards and snakes exhibit colour variation of adaptive value for thermoregulation, camouflage, predator avoidance, sexual selection, and speciation. Furcifer pardalis, the panther chameleon, is one of the most spectacular reptilian endemic species in Madagascar, with pronounced sexual dimorphism and exceptionally large intra-specific variation in male coloration. We perform here an integrative analysis of molecular phylogeography and colour variation after collecting high-resolution colour photographs and blood samples from 324 F. pardalis individuals in locations spanning the whole species distribution. First, mitochondrial and nuclear DNA sequence analyses uncover strong genetic structure among geographically-restricted haplogroups, revealing restricted gene flow among populations. Bayesian coalescent modelling suggests that most of the mitochondrial haplogroups could be considered as separate species. Second, using a supervised multiclass support vector machine approach on five anatomical components, we identify patterns in 3D colour space that efficiently predict assignment of male individuals to mitochondrial haplogroups. We converted the results of this analysis into a simple visual classification key that can assist trade managers to avoid local population over-harvesting.
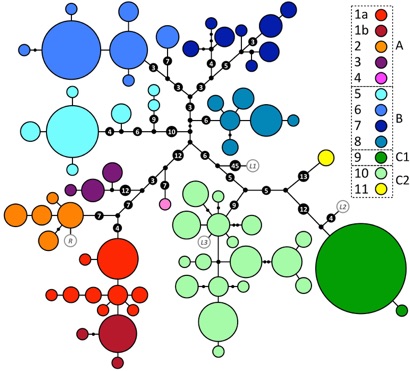
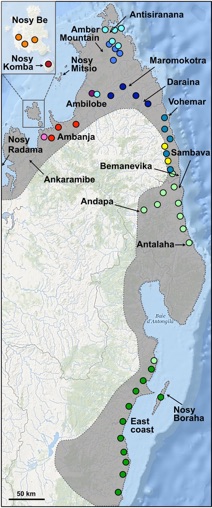
We reconstructed genealogical relationships among DNA sequences obtained from the 324 sampled panther chameleons. The figure below shows the mitochondrial DNA genealogical UMP network. Haplotypes are represented with coloured circles the sizes of which are proportional to the number of individuals. Inferred missing haplotypes are shown as small black dots and each line indicates a mutational change (numbers of mutations >1 are shown inside larger black dots). Sampling locations are indicated on the map below with colours corresponding to mitochondrial haplogroups shown in the genealogical network.
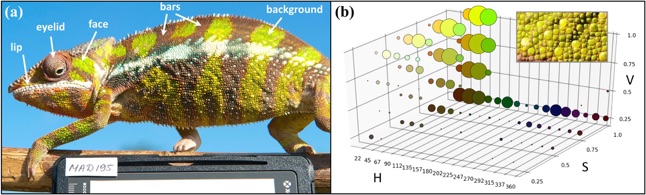
We then performed support vector machine (SVM) classification on the basis of colour characters extracted from five anatomy components (eyelid, lip, face, vertical bars and body background) in male panther chameleons. The top left image (a) in the figure above shows a male panther chameleon photographed with a white balance card; arrows indicate body parts used in colour analyses. The graph on the right (b) shows an example of an anatomical component (here background colour from the individual shown in (a)) and the corresponding 3D HSV histogram; the size of bubbles is shown on a logarithmic scale for ease of reading.
The trained classifier allowed for prediction of the position of many individuals in the mitochondrial genealogy (i.e., in either group A, B, C1, or C2) on the basis of their colour photographs. The results of this SVM analyses were converted into a simple classification key described in the original paper.
We aditionally performed species delimitation analyses using a Bayesian approach (Yang & Rannala 2010) on the combined nuclear and mtDNA dataset. These analyses suggested that the 11 haplogroups shown in the first figure above form 11 distinct species.
Discussion
The large number of animals sampled across the Malagasy panther chameleon range allowed us to reveal and characterise a greater degree of intra-population variability than previously reported. Our phylogeography and trained multiclass SVM classification of colour variation revealed strong genetic structure, with distant geographically-restricted lineages characterised by clade-specific male colour features (e.g., bright yellow lips in north-western haplogroups). Some of these lineages might warrant species status as suggested by our analyses based on a Bayesian approach to species delimitation. In case our results are confirmed, taxonomic revision would be warranted.
Additional analyses are also warranted to determine if the differences of colour characters among populations/haplogroups are associated to adaptation to local habitat and/or sexual selection. Note that, although panther chameleons from all geographical origin seem to successfully interbreed in captivity, anecdotal evidence (Ferguson et al. 2004) suggests that some of these hybrids present low reproductive performances, hinting at potential post-zygotic isolation and, hence, at the existence of multiple species of panther chameleons as suggested above.
High scores generated by the multiclass SVM indicate substantial predictive efficiency of the supervised machine learning classification approach. We suggest that these methods should be more widely used in phylogeography and classification studies because they can produce useful predictions when clustering approaches fail. Note that the key we devised is a vast simplification of the multidimensional decision function generated by the SVM and is therefore likely to be less performant than a full SVM analysis. However, the key being much more practical than SVM analyses, it could assist the Malagasy CITES Authority for managing the trade of wild-caught panther chameleons and avoid local population over-harvesting.
One limitation of our SVM analyses is that males might fully develop their coloration at a specific period of the year (cf. our observations of captive males), suggesting that the SVM results, and the corresponding classification key, might be best valid during the breeding season and/or in natural conditions.
We do not expect the ability of panther chameleons to change colour (through tuning of a 3D lattice of intracellular guanine nanocrystals (Teyssier et al. 2015)) to have affected our analysis as (i) panther chameleons actively change skin hue only during social interactions (such as when encountering a male competitor or a potentially receptive female), (ii) other colour changes, that can be induced by stress during capture, only affect skin brightness (i.e., diffuse and/or specular reflectivity) through dispersion/aggregation of melanosomes within dermal melanophores (Nilsson Skold et al. 2013; Taylor & Hadley 1970) and are therefore unlikely to affect our analyses, and (iii) colour photographs were taken on site immediately after capture, i.e., before potential reaction of the animal in terms of colour change.
Future work, investigating the physiology and genetic pathways involved in production of pigments and structural colours (Higdon et al. 2013; Ziegler 2003), is warranted in panther chameleons for a better understanding of the correlation between colour variation and the genealogical lineages (possibly species) we uncover here. In addition, the possibility to perform controlled captive breeding among individuals from different colour morphs, corresponding to different evolutionary lineages, opens the prospect of using gene mapping deep-sequencing approaches (Lamichhaney et al. 2012) to identify the genetic basis of adaptive colour variation in this spectacular lineage.
Publications
Please, consult the original publication below for references & much additional information.
✓Grbic D., Saenko S.V., Randriamoria T.M., Debry A., Raselimanana A.P. & M.C. Milinkovitch.
Phylogeography and Support Vector Machine Classification of Colour Variation in Panther Chameleons
Molecular Ecology (2015) 24, 3455–3466
 Additional File 1 (Additional Discussion, Tables S1-S4, Figs. S1-S6).
Additional File 1 (Additional Discussion, Tables S1-S4, Figs. S1-S6).
Coverage
Other related publications
✓Teyssier J., Saenko S.V., van der Marel D. & M.C. Milinkovitch.
Photonic Crystals Cause Active Colour Change in Chameleons
Nature Communications 6: 6368 (2015)
 Supporting Information (Suppl Figs 1-4, Suppl Table 1, Suppl Text and Refs)
Supporting Information (Suppl Figs 1-4, Suppl Table 1, Suppl Text and Refs)
Coverage
✓Saenko S., Teyssier J., van der Marel D. & M. C. Milinkovitch
Precise colocalization of interacting structural and pigmentary elements generates extensive color pattern variation in Phelsuma lizards
BMC Biology 2013, 11: 105