South-American dolphins (page 2)


South-American dolphins (page 2)



Click here to go back to page 1
RESULTS
Dusky Dolphins

Fig. 1. Median-joining network depicting the phylogenetic relationships among, and geographical assignment of, all dusky dolphin mtDNA haplotypes (filled coloured circles) based on cytochrome b DNA sequences: green, Peru; yellow, Argentina; red, South Africa. The size of each circle is proportional to the corresponding haplotype frequency. Missing intermediates are indicated by black circles except for the most central missing intermediate, noted by ‘CMI’. Each branch between two (sampled or missing) haplotypes indicates a single mutational step. The three underlined numbers indicate fixed substitutions separating Atlantic and Pacifc haplotypes. Arrows indicate alternative rooting positions (see full article for details).
A critical examination of network methods
During this work on dusky dolphin phylogeography, we analysed the sequence data with several widely used network estimation and rooting methods. The resulting intraspecific gene genealogies and rooting inferences exhibited substantial differences, underlying the limitations of some algorithms. Given that scientific hypotheses and management decisions depend strongly on inferred tree or network topologies, there is a clear need for a systematic comparative analysis of available methods. We performed such an analysis and developed of a new method for genealogical network inference. Check HERE for much additional information.
Extending the analysis to the nuclear genome & additional samples / localities
We then used nine nuclear species-specific microsatellite loci and two mitochondrial gene fragments (cytochrome b and control region), for investigating the processes that have shaped the geographical distribution of genetic diversity exhibited by contemporary dusky dolphin populations. A total of 221 individuals from four locations (Peru, Argentina, southern Africa, and New Zealand) were assayed, covering most of the species’ distribution range. Although our analyses identify a general demographic decline in the Peruvian dusky dolphin stock (recently affected by high natural and human-induced mortality levels), comparison between the different molecular markers hint at an ancient bottleneck that predates recent El Niño oscillations and human exploitation. Moreover, we find evidence of a difference in dispersal behaviour of dusky dolphins along the South American coast and across the Atlantic. While data in Peruvian and Argentine waters are best explained by male-specific gene flow between these two populations, our analyses suggest that dusky dolphins from Argentina and southern Africa recently separated from an ancestral Atlantic population and, since then, diverged without considerable gene flow. The inclusion of a few New Zealand samples further confirms the low levels of genetic differentiation among most dusky dolphin populations. Only the Peruvian dusky dolphin stock is highly differentiated, especially at mitochondrial loci, suggesting that major fluctuations in its population size have led to an increased rate of genetic drift.
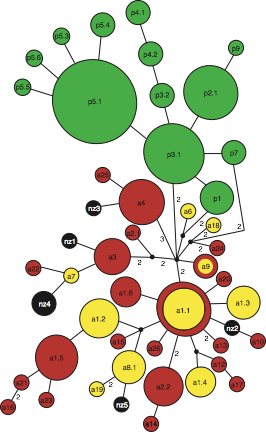
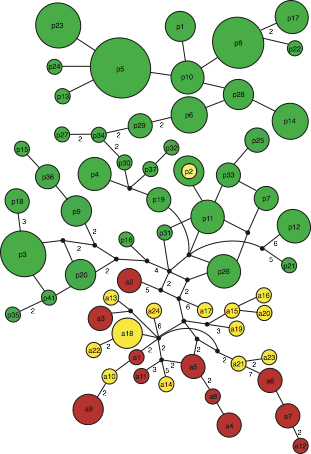
Fig. 2. Genealogical relationships among cytochrome b gene (a) and control region (b) haplotypes (green, Peru; yellow, Argentina; red, South Africa; black, New Zealand). Circle size is proportional to the number of individuals exhibiting the corresponding haplotype. Small unlabelled black circles represent missing node haplotypes. If not otherwise stated by numbers, a branch connecting two haplotypes corresponds to a single mutational step.
Check the full paper (Cassens et al. 2005, see below) for results on the statistics using the microsatellite makers.
Burmeister’s porpoises
Using mitochondrial control region sequences and 11 species-specific microsatellite loci, we assessed the genetic differentiation among 118 stranded, incidentally or directly-caught Burmeister’s porpoises from different localities in Peruvian, Chilean, and Argentine waters. F-statistics and Bayesian clustering analyses indicate a major population differentiation along the South American Pacific coast, separating Peruvian from both Chilean and Argentine individuals. Interestingly, this population boundary is consistent with the population structure found in the sympatrically-occurring dusky dolphin species (Lagenorhynchus obscurus; see results above). Given that vulnerability to local depletion for South American coastal porpoises and dolphins is probably highest in the Peruvian population (due to high exploitation levels and recurrent El Niño events), the genetic data reported here considerably strengthen the need for conservation efforts focused on regulation of catches in local waters. Our results hint at possible genetic differentiation among Burmeister’s porpoises (i) from the Atlantic and Pacific Ocean and (ii) from different Peruvian harbors. Finally, cross-species amplifications suggest that our newly-developed microsatellite markers will be useful in population genetic studies in the five other extant porpoise species.

Figure 3. Median-joining graph depicting the genealogical relationships among mitochondrial Burmeister’s porpoise haplotypes. Circle size is proportional to the number of individuals exhibiting the corresponding haplotype. Small unlabeled black squares represent missing, non-sampled haplotypes. Each branch connecting two haplotypes corresponds to a single mutational step with the dashed line representing the fixed substitution in the tRNA-thr gene that separates Peruvian and Chilean haplogroups.

The population boundary (Peru vs. other localities) observed both in Burmeister’s porpoises and dusky dolphins strongly suggest the presence of a cryptic barrier to gene flow (cf. figure on the left) whose nature remains to be identified.
“Forensis” analyses in fish markets
Despite a ban on small cetacean exploitation in 1990 and further reinforced in 1994, captures of several species, including dusky dolphins, Burmeister’s porpoises and long-beaked common dolphins (Delphinus capensis), still occur in Peruvian fisheries. Indeed, between 1995 and 1999, Van Waerebeek et al. reported on small cetacean exploitation in 83.5% of the visited harbors and found the remains of at least 452 captured cetaceans, 43.5% of which were P. spinipinnis. Given that hunting of small cetaceans is prohibited, fishermen increasingly try to commercialize their catches hidden from public view. As evidenced by beach-cast carcasses found in the proximity of harbors, cetacean specimens are increasingly cut up on board and parts of no value are discarded before entering the ports.
We have recently performed molecular identification of small cetacean samples collected in Peruvian fish markets. The results of these analyses are described HERE.
Publications
Please, consult the full publications below for references and much additional information.
✓Cassens I., Van Waerebeek K., Best P. B., Crespo E.A., Reyes J. & M. C. Milinkovitch
The phylogeography of dusky dolphins (Lagenorhynchus obscurus): a critical examination of network methods and rooting procedures
Molecular Ecology, 12: 1781-1792 (2003)
✓Cassens I., Van Waerebeek K., Best P.B., Tzika A., Van Helden A.L., Crespo E.A. & M. C. Milinkovitch
Evidence for male dispersal along the coasts but no migration in pelagic waters in dusky dolphins (Lagenorhynchus obscurus)
Molecular Ecology, 14 : 107-121 (2005)
✓Cassens I., Mardulyn P. & M. C. Milinkovitch
Evaluating Intraspecific “Network” Construction Methods Using Simulated Sequence Data: Do Existing Algorithms Outperform the Global Maximum Parsimony Approach?
Systematic Biology 54: 363-372 (2005)
✓Rosa S., Milinkovitch M.C., Van Waerebeek K., Berck J., Oporto J., Alfaro-Shigueto J., Van Bressem M.F., Goodall N. & I. Cassens
Population structure of nuclear and mitochondrial DNA variation among South American Burmeister’s porpoises (Phocoena spinipinnis)
Conservation Genetics 6: 431–443 (2005)
✓Mardulyn P., Cassens I. & M. C. Milinkovitch
A comparison of methods for constructing evolutionary networks from intraspecific DNA sequence
Pages 104-120 (Chapter 5) in ‘Population Genetics for Animal Conservation’ (Bertorelle G, Bruford M.W.,
Hauffe H.C., Rizzoli A., & Vernesi C., eds.),
Cambridge University Press 2009
✓Tzika A.C., D’Amico E., Alfaro-Shigueto J., Mangel J.C., Van Waerebeek K. & M.C. Milinkovitch
Molecular identification of small cetacean samples from Peruvian fish markets
Conservation Genetics 11: 2207-2218 (2010)





Back to the
“Conservation Genetics” page
Back to the
“LANE”
introduction page


FACTS
Species:
‣Lagenorhynchus obscurus
‣Phocoena spinipinnis
Locations:
‣Peru
‣Chile
‣Argentina
‣extended to South-Africa
& New Zealand

PUBLICATIONS
‣Molecular Ecology, 12: 1781-1792 (2003)
‣Molecular Ecology, 14 : 107-121 (2005)
‣Systematic Biology 54: 363-372 (2005)
‣Conservation Genetics 6: 431–443 (2005)
‣Pages 104-120 (Chapter 5) In “Population Genetics for Animal Conservation”, Cambridge University Press (2009)
‣Conservation Genetics 11: 2207-2218 (2010)

Related publications from our group on cetacean population genetics
‣Milinkovitch, LeDuc, Tiedemann, & Dizon
Applications of molecular data in cetacean taxonomy and population genetics with special emphasis on defining species boundaries. In: Marine Mammals: Biology and Conservation (eds Evans & Raga), pp. 325–359. Kluwer Academic/Plenum, New York (2002)
‣Tiedemann & Milinkovitch
Culture and Genetic Evolution in Whales
Science, 284: 2055a (1999)
‣Tiedemann., Hardy, Vekemans & Milinkovitch
Higher Impact of Female than Male Migration on Population Structure in Large Mammals
Molecular Ecology, 9: 1159-1163 (2000)

PEOPLE INVOLVED
FROM MICHEL
MILINKOVITCH’S LAB
‣Insa Cassens
‣Patrick Mardulyn
‣Sabrina Rosa
‣Athanasia Tzika
‣Jehanne Berck

MAIN COLLABORATORS
Koen Van Waerebeek
Peruvian Centre for Cetacean Research (CEPEC), Museo de Delfines, Pucusana, Lima, Peru
Julio Reyes
Areas Costeras y Recursos Marinos (ACOREMA), Pisco, Peru

Other collaborators and sample sources
‣Jorge Oporto
Corporacion Terra Australis, Avda. Alemania 630, Valdivia, Chile
‣ Joanna Alfaro-Shigueto & Jeffrey Mangel
Asociacion Pro Delphinus, Lima, Peru
‣ Marie-Francoise Van Bressem
Peruvian Centre for Cetacean Research (CEPEC), Museo de Delfines, Pucusana, Lima, Peru
‣Peter B. Best
Mammal Research Institute, University of Pretoria, South Africa
‣Enrique A. Crespo
Laboratorio de Mamiferos Marinos, Centro Nacional Patagonico, Puerto Madryn, Chubut, Argentina
‣Anton L. Van Helden
Museum of New Zealand Te Papa Tongarewa, Wellington, New Zealand

