Jamaican boas (page 2)


Jamaican boas (page 2)





Click here to go back to page 1
Molecular analysis of the Jamaican boa captive breeding program
Given the signs of decline of the natural population(s), a captive breeding programme was initiated in 1976 at the Durrell Wildlife Conservation Trust (Jersey, UK) and was rationalized in 2002 by incorporating pedigree information into a European Endangered species Programme (EEP) involving 14 member institutions of the European Association of Zoos and Aquaria (EAZA; www.eaza.net). Recently, using nuclear and mitochondrial DNA markers, we identified parental allocation errors and ambiguities in the EEP studbook, and assessed the genetic diversity and levels of inbreeding of the current captive population. Combining measures of relatedness derived from multilocus genotypes with practical parameters such as age of animals and localization of host institutions, we proposed a scheme of mating groups that would produce minimal inbreeding in the captive population of the Jamaican boa (Tzika et al. 2009).
History of the captive breeding program
Beside becoming increasingly involved in in-situ conservation projects, many zoos and aquaria around the world organize captive breeding programs to attempt generating self-sustaining populations for the long-term conservation of populations and species that are threatened or extinct in their natural habitat. For example, 165 species are incorporated into European Endangered species Programs (EEP) at member institutions of the European Association of Zoos and Aquaria (EAZA; www.eaza.net). The efficiency of these ex-situ conservation efforts is highly variable mostly because of the difficulties experienced by some species to breed in captivity (as exemplified by the giant panda, Ailuropoda melanoleuca, and the Sumatran rhinoceros, Rhinoceros sumatrensis) and/or the need to incorporate molecular genetic data for minimizing inbreeding and retaining population structure observed in the wild (see our work on the Giant tortoises from the Española Island in the Galápagos Archipelago). Here, we focus on the captive-breeding program of the endemic Jamaican boa (or ‘‘yellow boa’’, Epicrates subflavus), a spectacular species of the Caribbean biodiversity hotspot. The Jamaican boa remained abundant in its natural habitat from the arrival of the first Europeans in Jamaica in the 1500s until the end of the 19th century. Since then, natural populations of the Jamaican boa have greatly declined mainly due to predation by introduced species, human persecution, and habitat destruction (see our web page on the natural populations of the yellow boa). E. subflavus is listed today as a ‘vulnerable’ species by IUCN (Red List 2006). In response to increasing conservation concerns, a captive breeding program was initiated by the Durrell Wildlife Conservation Trust (DWCT; Jersey, UK).
In 1976, seven wild-born individuals (four males and three females) were transferred from Hope Zoo, Kingston, to Europe; subsequently, an additional 4 wild-born individuals (three females and one male) were added to the group of ‘‘founders’’ in 1993 (see first Figure below). During the last 30 years, more than 600 offspring have been produced and distributed among European zoological institutes and private facilities/individuals. In an effort to rationalize the breeding program, pedigree information on the captive population started to be incorporated in 2002 into an EEP-EAZA studbook. Information gathered from systematic questioning of institutions and private individuals known to house ‘‘founders’’ and/or their descendants complemented the data already available on ISIS. Not surprisingly, the process did not generate a comprehensive studbook because (i) the exact geographic origin of, and genetic relatedness among, the ‘‘founders’’ were unknown, and (ii) many institutions and private individuals did not keep accurate records. Nonetheless, it has been possible to deduce many of the transfers of animals that occurred as well as to reconstruct a significant portion of the pedigree with reasonable confidence. Finally, it should be noted that peculiarities in the mating behaviour of the species (reproduction is facilitated by putting at least two males in the presence of a single female) generated additional ambiguities in the pedigree.
Here, using nuclear and mitochondrial molecular markers, we:
(i)assess the genetic diversity of the current captive population of the Jamaican boa,
(ii)perform deterministic parental assignment of each sampled offspring,
(iii)compute the reproductive success of each breeding individual,
(iv)estimate the effective population size (Ne) of the captive population, and
(v)suggest optimal mating groups for maximizing diversity.
When put into perspective with the genetic variability and structure of the surviving Jamaican natural populations, the data presented here allow us to (i) determine the origin of the captive population founders, and (ii) estimate the proportion of the species diversity preserved in the captive population. Our analyses provide guidance for better management of the ex-situ population, highlight the need for proper management of in-situ captive subpopulations, and might facilitate potential future repatriation programs.
Results: (1) levels of genetic diversity and inbreeding
We constructed a microsatellite-enriched genomic DNA library of E. subflavus and all sampled individuals were typed for nine microsatellite loci using three multiplex-PCR reactions (see full publication for details). Using a previously available mitochondrial cytochrome b sequence of E. subflavus, we designed primers for the amplification of a 647-bp fragment. Parental allocation, relatedness, and genetic variability statistics were assessed using the softwares “Pasos”, “Kinship v.1.3.1”, “Convert”, and “Fstat v.2.9.3”.
Note: We computed effective population sizes of females and males taking into account the observed variance in reproductive success. The effective population size is the number of breeding individuals in an idealized population that would show the same amount of inbreeding as the population under consideration. When there is a biased sex ratio, the effective population size is reduced following:
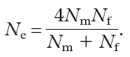
where Nf and Nm are census numbers of females and males, respectively.
The effective population size is further reduced if different individuals exhibit different reproductive success. In that case, Ne of females and males (Nef & Nem) can be computed as follows:
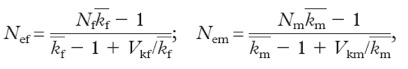
where kf and km are the means of offspring number per female and male breeder, respectively, and Vkf and Vkm are the variances in offspring number per female and male breeder, respectively.
Among the 601 individuals known to belong to the captive population, 162 offspring cannot be located or tracked (many of them probably died and were disposed of without the event being recorded), 170 died in their first year of captivity, 184 died at a later stage, and 85 surviving individuals (founders and offspring) are currently distributed at 14 European institutions (Fig. 1). Among the F0 individuals, three males and four females have since died. All of the 47 sampled individuals (numbers with rectangular frame in Fig. 1) were successfully sequenced for the target cytochrome b fragment and successfully genotyped for the nine selected polymorphic microsatellite loci.
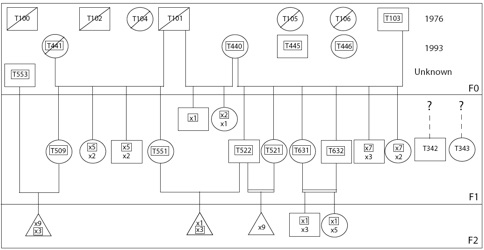
not reproduce. Double horizontal lines depict pairs of siblings, whereas dashed lines indicate the unknown position of the corresponding individuals in the pedigree. Framed numbers correspond to genotyped individuals. Dead individuals (before May 2007) areindicated by a diagonal stripe
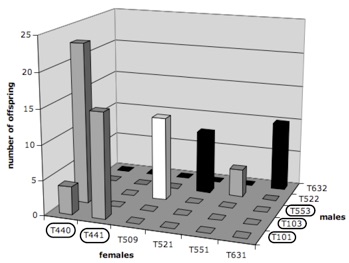
Using the molecule data, we estimated the percentage of uncollected parents in the first (founding) generation to 3% of uncollected females and 33% of uncollected males. These numbers are in good agreement with information available in the studbook: all living offspring of the first generation are likely to originate from two female (T440 & T441) and two male (T101 & T103) founders, with T101 who died early in the breeding program (hence, has not been sampled). In other words, as far as we know, other F0 individuals (of which two, T445 and T446, are still alive) have not contributed offspring to the breeding program. By comparing the offspring-mother genotypes of all F1 individuals sired by the uncollected male founder (T101), we could reconstruct the genotype of the latter. Both parents can be deterministically assigned to each genotyped offspring in the captive population except one individual (T553; Fig. 1) that exhibits three alleles not found in the sampled founders. We therefore assume that T553 is either the offspring of dead founders or was wild-born and, as such, is included in the F0 group for further analyses (Fig. 1). Two F1 adults (one male, T342, and one female, T343) could not be sampled and have an unknown history in the pedigree (Fig. 1).
Parental assignment based on molecular data also shows that studbook information was erroneous for eight (24%) of the 33 sampled F1 individuals. A closer look at birth dates and locations as well as at the sizes of the litters to which the problematic individuals belong, indicates that there probably was a mix up among three litters of animals born at the same place within a one-month interval. Furthermore, the molecular analyses also clarify the sire identity for 11 (33%) of the 33 sampled F1 individuals, an uncertainty that originated from the presence of multiple males in association with a single female for mating facilitation.
The distribution of the cytochrome b haplotypes generated here indicates that all F1 and F2 individuals originate from two maternal lineages that correspond to the two sampled F0 females (T440 and T441, whose haplotypes are separated by three mutations). Maternal allocations of all sampled offspring inferred by the mitochondrial marker confirm the ones based on the microsatellite genotypes.

Results: (2) managing the captive breeding program
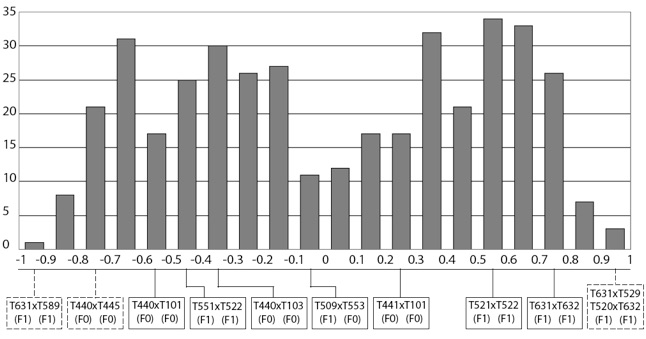
As we here disregard selection, loss of diversity in the captive population can only be brought about by inbreeding and genetic drift which are basically two different ways of looking at the same phenomenon (both drift and inbreeding increase mean relatedness among individuals). Hence, estimates of relatedness, computed from multilocus microsatellite genotypes is a particularly important statistic for devising a mating structure that would best achieve the goal of maximizing diversity of the captive population. The figure below (showing the distribution of pairwise genetic relatedness values, computed from microsatellite data, for all possible [female x male] pairs; Dashed frames indicate putative pairs whereas plain frames indicate actual pairs, i.e., that already produced offspring) shows the distribution of pairwise genetic relatedness for all possible [female x male] combinations (regardless of the age of the individuals) based on our microsatellite data. According to our analysis, the two worse mating pairs would be [T631 x T529] and [T520 x T632], whose members are all offspring of [T440 x T103]. The worst pairs of actual mates are the [T631 x T632] and [T521 x T522] pair of siblings (R = 0.79 and 0.60, respectively). Conversely, the putative best mates (i.e., with lowest relatedness) would be a pair of F1 offspring [T631 x T589] with R = -0.94, whereas the best mating pair among F0 individuals would be [T440 x T445] with R = -0.72. Among the actual mates (i.e., those that did produce offspring in the captive breeding program so far), relatedness values range from -0.57 to 0.79, with the lowest values attributed to a pair of F0 individuals [T440 x T101]. Note however that this ‘best actual mating pair’ of individuals only ranks 68th in the list of 399 putative mating pairs sorted by increasing relatedness (Fig. below). In other words, 67 (17%) of the other possible [female x male] pairs would generate more diversity in the captive population.
However, beside genetic relatedness, other parameters such as the age of the individuals, the localization of host institution, and the need to group more than two animals for inducing mating, are also relevant to the design of an efficient breeding program. We propose mating groups in the table below. In each such group, at least two mature males and one mature female are included, and any possible corresponding male-female pair exhibits a low relatedness (whereas animals of the same sex may exhibit higher relatedness values). Note that the best possible mating pair [T631 x T589] is not formed, as a result of an effort to minimize movements among host institutions. We also suggest that littermates born recently (hence, of unknown sex and with a high mortality rate) are distributed among host institutions to avoid inbreeding if they survive. The average of the mean male-female relatedness values within the groups listed in the table is -0.36, i.e., a value much better (lower) than the average relatedness among all possible pairs (0). Of course, other parameters, such as nonrandom mating (caused by, e.g., social dominance among males) and differences of fertility among males and among females will also need to be taken into account for possible future modifications of the captive breeding program.
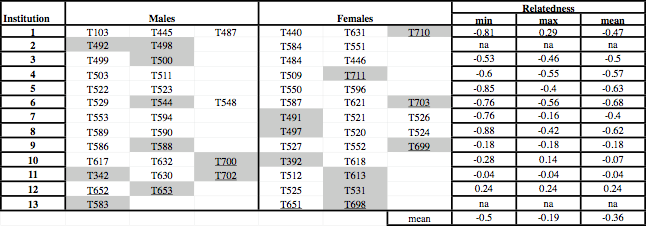
Table: Suggested new mating groups among Jamaican boas for the captive breeding program. Groups were designed by minimizing movements among host institutions and relatedness values of each possible female/male pair within groups. The identification numbers of unsampled individuals are shaded and of sub-adults (< 6 years old) are underlined. The minimum, maximum, and mean relatedness values within each group are given (na, not applicable).
Finally, Cytochrome b haplotype network analyses indicate that all sampled F0 individuals originate from one of the three natural populations still surviving in Jamaica (see page 1). Hence, the current captive breeding program did not disrupt the gene pool of well-differentiated populations. However, if the captive breeding program aims at retaining both diversity and population structure, new individuals will need to be captured in the wild for (i) establishing an extended breeding program incorporating all natural populations and (ii) rectify the loss of allelic diversity in the current captive population (whose allelelic richness is almost half of that observed in the corresponding natural population, see full papers).
Conclusions and utility of this study
Detailed studbooks constitute the simplest means for proper management of captive populations. However, correct parental allocation of individuals is not always possible without the use of molecular data, and pedigree information is not necessarily sufficient for the selection of the best breeding pairs (i.e., least related individuals). Decisions based on erroneous or incomplete data may lead to decrease of genetic diversity and loss of genetic structure, hence, can jeopardize the long-term survival of captive and/or repatriated populations. Inbreeding is probably best avoided in captive breeding and restauration programs by maximizing variability and retaining the population structure observed in the wild. This is the approach we use here (combination of studbook data and molecular genetic analyses), focusing on the captive breeding program of the endemic and vulnerable Jamaican boa, Epicrates subflavus. Many of the captive individuals, including those founders still available to sampling, were sequenced at a fragment of the mitochondrial cytochrome b gene and genotyped at nine nuclear microsatellite loci. Analyses of these molecular data pointed out to parental allocation errors and clarified ambiguous sire assignments. Despite that the observed and expected heterozygosity values of the F2 generation are not significantly lower than those of the founder population or of the wild population from which, the founders originated (see full publication), a decrease in variability is clearly detected by the loss of alleles (Fig. 3) and the significant difference of allelic richness (see full publication) between the F0 and the F2 generations. This illustrates the limited efficiency of tests based on heterozygosity variations to detect recent inbreeding.
During the last 30 years, the Jamaican boa European captive breeding program generated more than 600 offspring, of which 80 are still alive/tracked today. Note that specimens are also available in North-American zoos; although it is possible that some of these individuals were derived from DWCT founders, further analyses are warranted to evaluate the possible value of incorporating these animals into the European breeding program. Important modifications are required for the maintenance or strengthening of the captive population genetic diversity. Indeed, although a total of 12 wild animals were incorporated into the program, only the offspring of four of them are still alive today. Furthermore, (i) many of the sexually-mature offspring of first and second generation never reproduced, and (ii) more than half of the offspring of second generation have full siblings as parents (see above).
Variance in reproductive success generated a drop of Ne, the effective population size, from 58.6 to 8.69. This low Ne value raises the specter that severe inbreeding depression and reduced viability/adaptability are possible. Thus, if an ex-situ breeding program is being carried out with the hope to serve for a repatriation program into the restored natural habitat of the species, the probability of species long-term survival will almost certainly be increased by efforts to restore as much genetic variation as possible in the captive population both by minimizing relatedness among breeders and by the incorporation of additional wild-caught individuals from the corresponding natural population.
Note however that using a captive-bred population as the source for repatriation programs is not without drawbacks. For example, captive-bred animals can bear alleles generating low fitness in the wild and repatriation raises the risk of introducing non-native pathogens and parasites. Hence, some conservationists would argue that a better approach would be to develop an in situ captive breeding program (as carried out in the Galápagos for giant tortoises of the Española island). We demonstrate here that the use of molecular data has the potential to improve management of the captive breeding program well beyond what can be achieved with pedigree information alone. Indeed, analyses of the molecular data allowed
(i)identification of the natural population from which the founders originated, hence, avoiding to disrupt population structure,
(ii)detection of errors/ambiguities in the pedigree, hence, avoiding possible mating of closely related individuals,
(iii)computation of relatedness values among individuals for the design of optimal mating groups (i.e., an improvement over equalization of founder contributions).
Furthermore, molecular data would allow the identification of best candidates for introduction of new wild-born individuals in the captive breeding program. For example, our analyses indicate that the two F0 individuals (T445 & T446; Fig. 1) which did not yet contribute any offspring (i) originate from the same natural population as the other founders, and (ii) exhibit 2 alleles (at 2 loci) not observed in the genotypes of the successful breeders. The low relatedness values between, on one hand, any of these two individuals and, on the other hand, successful F0 or F1 breeders further support the conclusion that these animals should indeed be integrated.
None of the above-mentioned goals is feasible without the active collaboration of the multiple institutions hosting Jamaican boas. Such an attempt may face numerous difficulties and will require modifications of common procedures. For example, when an institution requests the acquisition of new specimens, it is a regular practice to provide it with several littermates, a few of whom will survive to adulthood. Indeed, members of the zoo personnel are usually reluctant to give away mature animals for which they cared for years. The usual final result is that siblings reproduce with each other in the new hosting institution. Ideally, there should exist a single institution hosting and breeding all the animals incorporated in the breeding program. However, such a situation is difficult to reach because of the limited financial and space capacities of most zoos and because it would conflict with another important goal: incorporate a sufficiently large number of institutions in the ex-situ conservation program of the Jamaican boa for promoting public awareness through their exhibitions. Combining theoretical parameters based on molecular data (such as relatedness values of mating pairs based on multilocus microsatellite genotypes) and practical parameters such as age of animals and localization of host institutions, we propose mating groups (see Table above) that would maximize diversity in the captive breeding program of the Jamaican boa.
✓Tzika A. C., Koenig S., Miller R., Garcia G., Remy C. & M. C. Milinkovitch
Population structure of an endemic vulnerable species, the Jamaican boa (Epicrates subflavus)
Molecular Ecology 17, 533-544 (2008)
✓Tzika A. C., Remy C., Gibson R. & M. C. Milinkovitch
Molecular Genetic Analysis of a Captive-Breeding Program: The Vulnerable Endemic Jamaican Yellow Boa
Conservation Genetics 10:69–77 (2009)

Back to the
“Conservation Genetics” page
Back to the
“LANE”
introduction page


FACTS
Species:
‣Epicrates subflavus
Locations:
‣Jamaica

PUBLICATIONS
‣Molecular Ecology 17, 533-544 (2008)
‣Conservation Genetics 10:69–77 (2009)

Related publications from our group on population genetics
‣Check the main “Conservation Genetics” page

PEOPLE INVOLVED
FROM MICHEL
MILINKOVITCH’S LAB
‣Athanasia Tzika

MAIN COLLABORATORS IN JAMAICA
The Windsor Research Center
was established to promote, facilitate, monitor and centralise research associated with the Cockpit Country. The center is run by:
✓Susan Koenig
✓Michael Schwartz
who where both pivotal in providing their logistic and scientific expertise.
NEPA (National Environment and Planning Agency)
The following people at NEPA have provided their technical & logistic expertise:
✓Yvettes Strong (head of the "Biodiversity Branch")
✓Ricardo Miller
✓Canute Tyndale
✓Andrea Donaldson

THE CAPTIVE BREEDING TEAM
The captive breeding program was initiated in the 1970s at the Durrell Wildlife Conservation Trust (Jersey Zoo, UK).
The captive breeding was rationalized in 2002 by incorporating pedigree information into a European Endangered species Programme (EEP) involving 14 member institutions of the European Association of Zoos and Aquaria (EAZA). The coordinator of this EEP is:
✓Christophe Remy (vivarium Tournai, Belgium)
The 14 member institutions involved in the captive breeding programme of the Jamaican boa are:
✓The Musée d’Histoire Naturelle de Tournai (Belgium)
✓The Birmingham Nature Center (UK)
✓The Blackpool Zoo (UK),
✓The Broxbourne Paradise Wildlife Park (UK)
✓The Cotswold Wildlife Park (UK)
✓The Chessington World of Adventures (UK)
✓The Chester Zoo (UK)
✓The Durrell Wildlife Conservation Trust (Jersey, UK)
✓The Zoologicka Zohrada Jihlava (Czech Republic)
✓The Lisboa Jardim Zoologico (Portugal)
✓The Lodz Miejski Ogrod Zoologiczny (Poland)
✓The Zoological Society of London (UK)
✓The Tula Exotarium (Federation of Russia)


LINKS

