large mammal populations






Back to the
“Conservation Genetics” page
Back to the
“LANE”
introduction page

FACTS
Species:
‣Large matriarchal mammals such as some whale species & elephants
Locations:
‣Inside the computer ;-)

PUBLICATIONS
‣Science, 284: 2055a (1999)
‣Molecular Ecology, 9: 1159-1163 (2000)

Related publications from our group on population genetics
‣Check the main “Conservation Genetics” page

PEOPLE INVOLVED
FROM MICHEL
MILINKOVITCH’S LAB
Ralph Tiedemann
Ralph was a Post-doc in Michel Milinkovitch’s lab. He is now Full Professor at the University of Potsdam, Germany.

OTHER PEOPLE INVOLVED
Olivier Hardy
was a PhD student in Xavier’s Vekemans’ laboratory at the time of the study. He is now group leader in the laboratory of Evolutionary Eco-Ethology at the University of Brussels (Belgium)
Xavier Vekemans
was a group leader in the Plant Biology department at the University of Brussels (Belgium). He is now Professor at the Laboratory of Genetics and Evolution of plant populations at the University of Lille 1 (France)




An appealing hypothesis of genetic hitchhiking caused by cultural traits
Hal Whitehead noted [Science 282, 1708 (1998)] that mtDNA variation is about tenfold lower in matrilineal whales than in whales without that specific social structure. He interpreted this correlation as selection of maternally transmitted cultural traits on which neutral mtDNA alleles “hitchhike.” His hypothesis requires that selection of cultural traits occurs in whales, as well as the questionable additional assumption that lateral transmission of the behavior to unrelated females is below 0.5%.
We however suggest [Science, 284: 2055a (1999)], rather, that lower variability of mtDNA in matrilineal whales does not require selection of mtDNA haplotypes or linked cultural traits because any stochastic heterogeneity in fecundity through space and time will cause a drastic reduction of mtDNA variability in matrilineal populations. To test our hypothesis, we used sperm whale life history parameters and simulated the effect of (i) matrilineal social structure and (ii) stochastic heterogeneity in fecundity on mtDNA variation.
Our results show that a large drop in mtDNA diversity occurred only when matrilineal structure was
implemented (Fig. 1). Stochastic differences in reproduction through time and space increase the variance in reproductive success among haplotype matrilines -- causing a decrease in mtDNA variation observed in the whole population -- only if these haplotype matrilines are spatially associated through the existence of individual matrilines (that is, matrilineal social structure). It is unlikely that the environment will be so homogeneous as not to contribute to variation in reproductive success through evolutionary time and across the whole range of the species distribution. At any time, variations in the environment will slightly increase the reproductive success of animals in some areas as compared with others. However, this heterogeneity will cause rapid extinction of some haplotypic matrilines, while others will flourish, only if individuals bearing haplotypes from the same mitochondrial lineage are spatially associated; that is, if their social organization is matrilineal. This principle holds even if matrilineal groups migrate a great deal.
As simulated here, the heterogeneity in reproductive success is stochastic in the sense that specific mtDNA haplotypes and cultural traits can be perfectly neutral in respect to variations in fecundity of individuals. Even if one would assume perfect neutrality of all characters (morphological, molecular, and behavioral), matrilineality itself is sufficient to cause low genetic variability when the environment (thus, the fecundity) is heterogeneous through time and space. Our hypothesis does not require any character that produces differential fitness to be transmitted from one generation to the next. It fits the data in Whitehead’s report more closely and parsimoniously than does the idea of hitchhiking of neutral mtDNA alleles through selection on maternally transmitted cultural traits.

Higher impact of female than male migration on population structure
Migration is an evolutionary force of great importance in population stratification. In mammals, sex-biased migration rates are common: one sex is often philopatric (i.e. faithful to a reproductive site), while the other is more dispersing. Gender-specific migration rates, and their impact on population structure, can be investigated through molecular genetic techniques, in particular by comparing mitochondrial (mt) to Y-chromosomal, and/or autosomal data. To investigate the impact of sex-specific migration rates on population structure in large mammals, we developed an individual-based stochastic population model which implements some of their specific life history characteristics, such as largely overlapping generations, slow reproduction, and extensive female parental care. In this paper, we use our model to investigate, through simulations, the genetic effects of various sex-specific migration rates under isolation-by- distance, using explicit life-history parameters from two illustrative large mammalian species: the blue whale (Balaenoptera musculus) and the Asian elephant (Elephas maximus).
Data analysis
According to the neighbourhood size concept of gene flow, the degree of spatial differentiation in a continuous population depends on the average migration distance of genes. We estimated gene migration for each sex separately as the distance (averaged across all individuals throughout each simulation run) between the position where an individual is conceived and the position where the father (for male gene migration) or the mother (for female gene migration) was conceived. Average gene migration (σgene) is the mean between male and female gene migration. For a linear habitat, this translates into neighbourhood size (N) with N= 2σgene*d*√π, where d is the density per distance unit. Since d = 1/3 in our case (1000 individuals per 3000 units), the neighbourhood size is given by N≈1.18*σgene. The smaller σgene (and, in consequence, N), the more spatial genetic structure develops in a population. To further illustrate this relationship with a measure widely used in molecular studies on real populations, we calculated fixation indices (FST) after arbitrarily subdividing the virtual line at the end of each run into three subregions of equal size (FST-NC, based on nuclear expected heterozygosity; FST-MT, based on mitochondrial haplotypic diversity).
Results and discussion
The effects of varying either male or female migration rates on spatial genetic structure are illustrated in Fig. 1. At the nuclear locus, approximately twice as much male than female migration is required to yield the same value of gene migration (σgene, Fig. 1a). This latter parameter is the primary determinant of spatial population structure, as confirmed by the highly significant negative correlation between σgene and FST-NC: r= –0.99 for Balaenoptera, r= –0.98 for Elephas. For each couple of symmetric combinations in which one of the two sexes disperses more than the other, lower FST values are obtained when females rather than males are the dispersing sex (Fig. 1b). Hence, our results surprisingly suggest that a given increase in female migration impacts the population genetic structure at nuclear loci (i.e., FST-NC) significantly more than the same increase in male migration (Fig. 1b). We suggest that the primary factor responsible for that unexpected finding is ‘female/offspring comigration’, i.e. the fact that each offspring comigrates with its mother for the time of gravidity and lactation. Hence, female migration impacts not only females, but also the comigrating offspring regardless of its sex. This causes a higher impact of female than male migration on the average gene migration (σgene) and — in consequence — on genetic population structure.
If our hypothesis is correct, the effect should: (i) disappear when gravidity and lactation are absent (i.e., set to 0 in control simulations); and (ii) increase with an increased length of gravidity and lactation. Expectation (i) was confirmed by control runs (data not shown), while expectation (ii) was met as we increased the duration of female/offspring comigration from two years (blue whale parameters) to four years (elephant parameters).
Because mitochondrial DNA (mtDNA) is maternally inherited, genetic structure at that locus (e.g., as estimated from FST-MT values) is generally expected to be solely influenced by female migration. However, our simulations show that male migration has a small but statistically significant influence on the degree of geographical structure of the population when FST-MT is calculated from the whole population (Fig. 1c). We show that this result simply originates from the inclusion of male migrants in the calculations. This suggests that, when male migrants have a high probability to be sampled (e.g., when male migration and/or life-expectancy are/is high), spatial genetic structure at the mtDNA should be assessed by investigating females only (Fig. 1d).
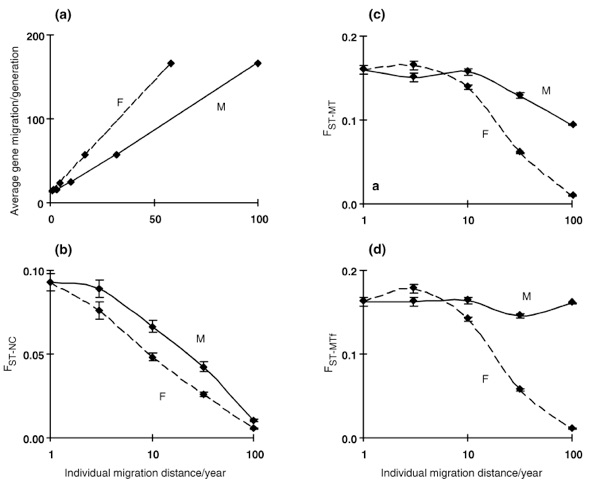
Fig. 1 Effects of increasing migration (measured with σ, the standard deviation in migration distances) of females (dotted line; σmale set to the fixed value 1) and males (plain line; σfemale set to the fixed value 1) on estimators of population structure. (a) Average gene migration for nuclear genes (σgene). Note that σgene translates into neighbourhood size (N) according to N ≈ 1.18σgene (see methods section in full publication). (b) Fixation index for nuclear genes (FST-NC). (c) Fixation index for mt genes, calculated from all individuals (FST-MT). (d) Fixation index for mt genes, calculated from females only (FST-MTf). Data points are medians (100 replicates) with standard errors where applicable. The difference in FST for σ = 3 [(c) and (d)] is statistically not significant. Life-history parameters are from the Asian elephant.
Although we only considered the special case of large mammals where ‘female/offspring comigration’ is extensive because of long gravidity and lactation periods, a similar effect should be observed in any species where mother and offspring comigrate for a significant time relative to generation time. Furthermore, species with male ‘pregnancy’ (e.g., pipefishes and seahorses) should exhibit a mirror-image effect with predominance of male rather than female migration. We suggest that the effect of ‘sex-specific dominance’ should be accounted for when spatial genetic structure of large mammal populations is inferred from nuclear molecular markers such as nuclear microsatellites. Furthermore, the concept of ‘strong-migration limit’ for geographically structured populations indicates that, when the level of migration exceeds a certain threshold relative to the effective population size (Ne), it dominates other evolutionary forces which might otherwise produce significant population subdivisions. When ‘mother/offspring comigration’ occurs, we suggest that this ‘strong-migration limit’ will be reached faster when female migration, rather than male migration, is increased
Please, consult the full publications below for references and much additional information (including stochastic model description):
✓Tiedemann R. & M. C. Milinkovitch
Culture and Genetic Evolution in Whales
Science, 284: 2055a (1999)
✓Tiedemann R., Hardy O., Vekemans X. & M. C. Milinkovitch
Higher Impact of Female than Male Migration on Population Structure in Large Mammals
Molecular Ecology, 9: 1159-1163 (2000)
